The structural basis for cohesin-CTF-anchored loops
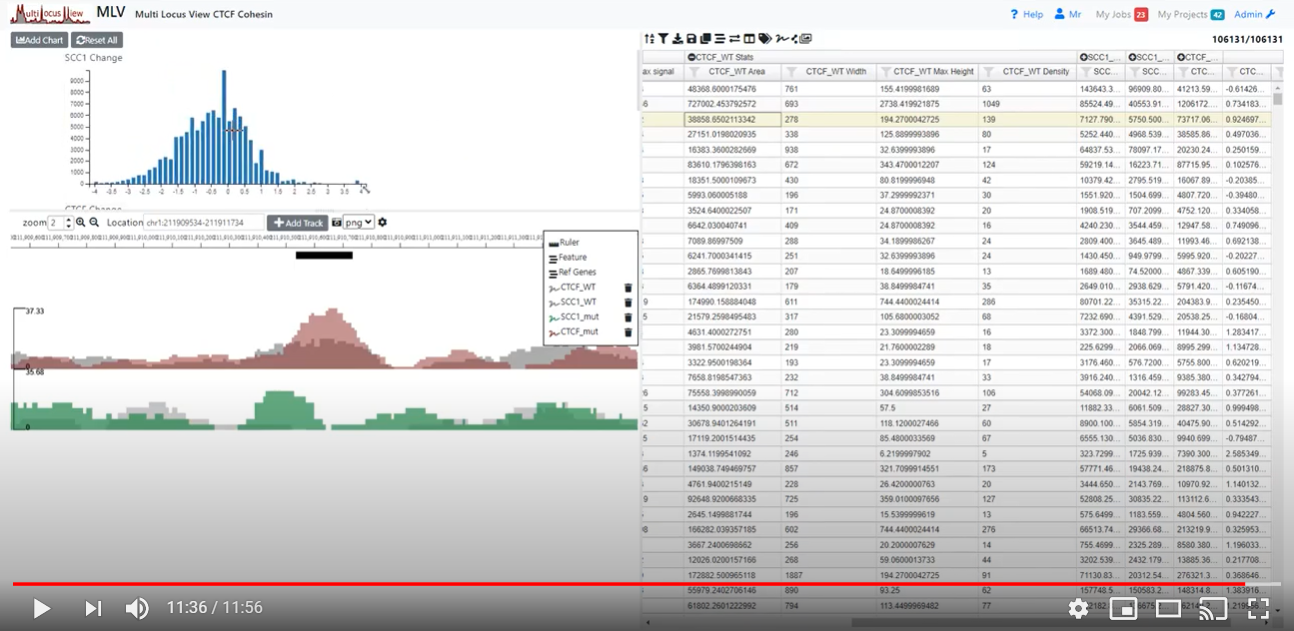
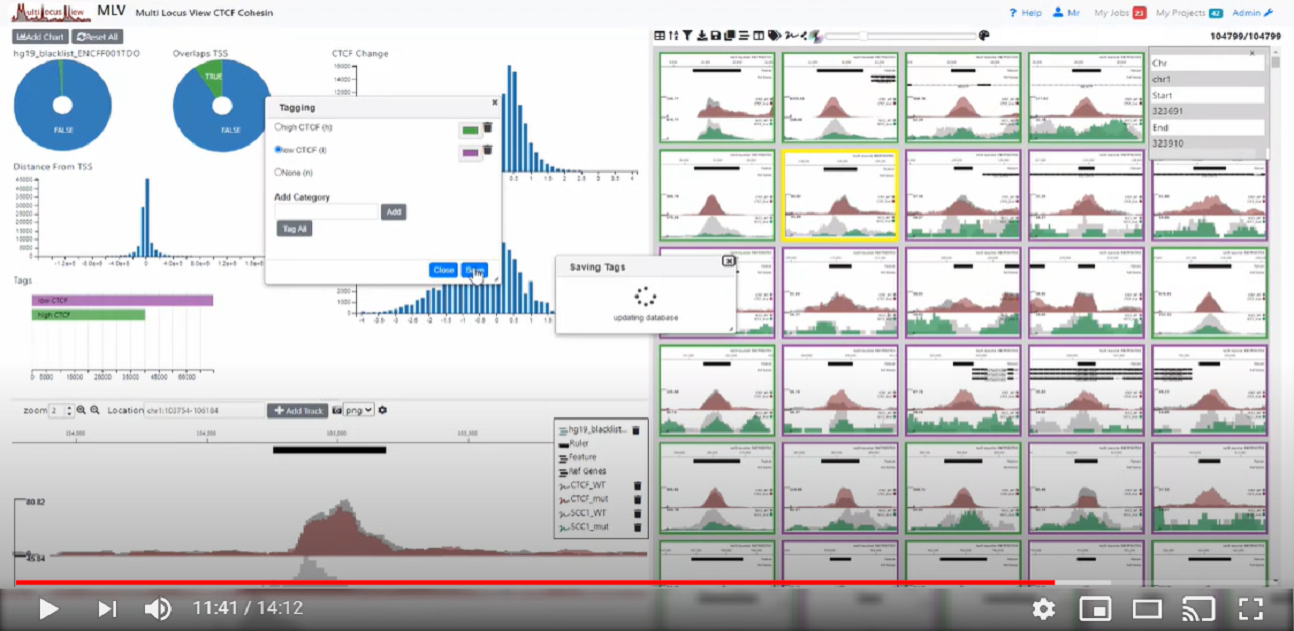
Tracking and understanding data quality, analysis and reproducibility are critical concerns in the biological sciences. This is especially true in genomics where Next Generation Sequencing (NGS) based technologies such as ChIP-seq, RNA-seq and ATAC-seq are generating a flood of genome-scale data. These data-types are extremely high level and complex with single experiments capable of mapping 10-100’s of thousands of biologically meaningful events across the genome. However, such data are usually processed with automated pipelines resulting in tabular outputs, which are difficult to verify and interpret without looking at the underlying data and combining it with data from other experiments. Conventional genome browsers are limited to single locations and do not allow for interactions with the dataset as a whole. MLV has been developed to allow users to fluidly interact with genomics datasets at multiple scales, from complete metadata labelled and clustered populations to detailed representations of individual elements. It has inbuilt tools to integrate signals across multiple dataset and to perform dimensionality reduction and clustering analysis.
Watch a video demonstrating the basic functionality of MLV
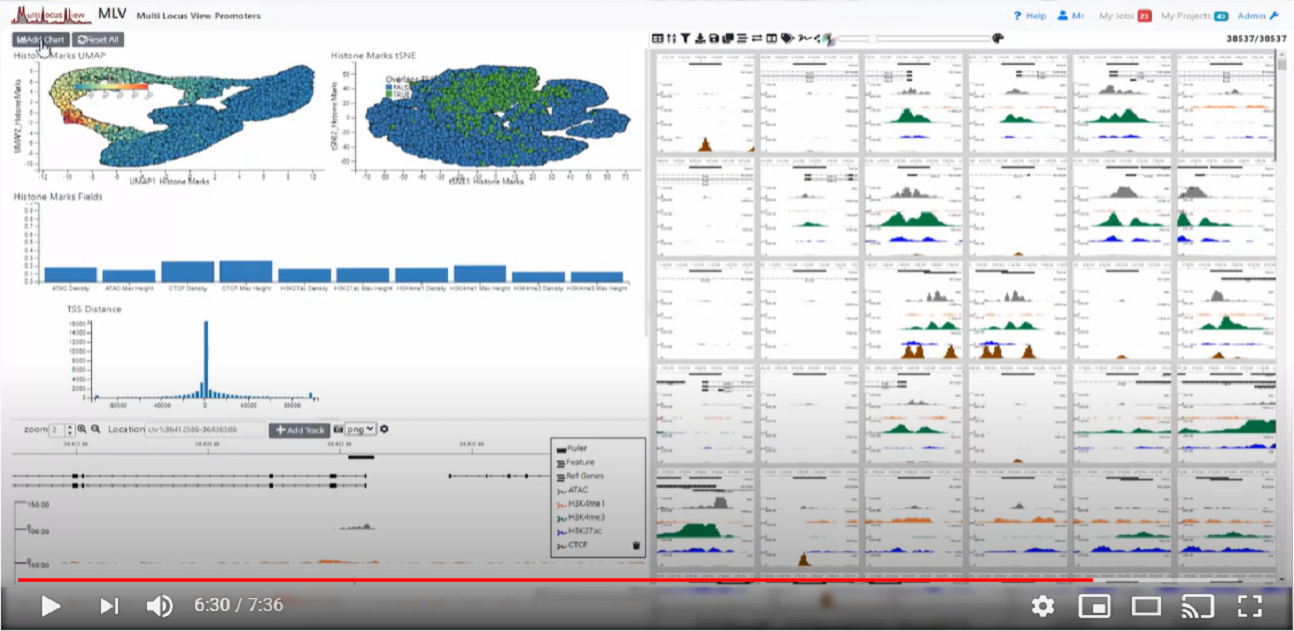

MLV is designed to be modular and allows development of other NGS visualisation solutions